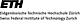plprot
- Plastids from light grown Arabidopsis ["chloroplast"]

|
Further details in:
Kleffmann et al., Current Biology 14, 354-362
Baginsky et al., Journal of Proteome Research 4, 637-640
|
Chloroplasts develop and differentiate from proplastids in photosynthetic tissues
in the presence of light. They are of central importance for the cellular metabolism
and play numerous unique roles in processes of global significance, e.g. photosynthesis
and amino acid biosynthesis. Chloroplasts are of cyanobacterial origin, but during
evolution they lost their autonomy and transferred most of their genes to the nucleus
(Martin and Herrmann, 1998). Since chloroplasts perform such important functions as
e.g. the synthesis of amino acids, fatty acids, carbohydrates and tetrapyrroles their
proteome has been studied in detail (reviwed in Baginsky and Gruissem, 2004,
van Wijk, 2004). Chloroplasts are structurally characterized by a sophisticated
internal membrane system, the thylakoid system, that is the site of photosynthetic
electron transport.
For our analysis, we have isolated chloroplasts from 7 week old short day grown
Arabidopsis plants by Percoll density gradient centrifugation. Following a serial
extraction of proteins based on solubility, we devised a multidimensional protein
fractionation strategy (FIGURE 1). All proteome analyses reported to date are
hampered by the dominance of photosynthetic proteins, that make the detection of
low abundance proteins difficult, if not impossible. We therefore decided to
deliberately enrich proteins from the chloroplast envelope membrane system to
identify those proteins that play a role in the communication of the chloroplast
with the cytosol (FIGURE 1). We identified a number of proteins that were not
previously predicted to localize to the chloroplast and are currently in the
process of establishing the chloroplast localization of a subset of these proteins
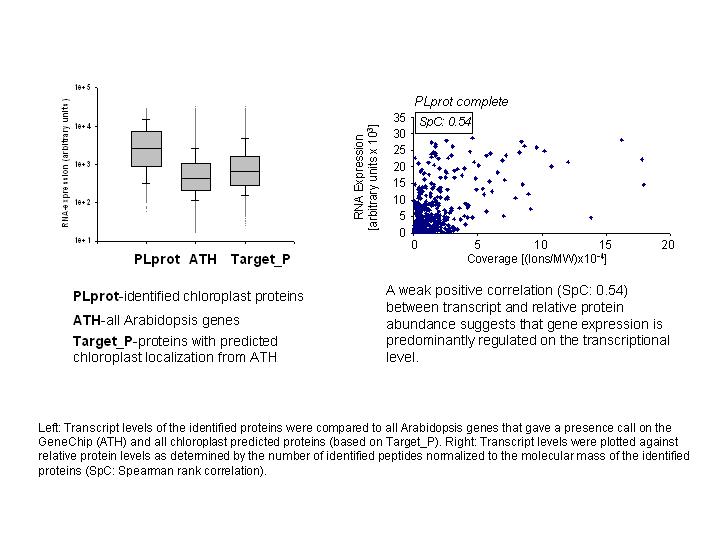
by additional analyses. Parallel transcriptional profiling suggested, that up to
date we only identified the most abundant proteins, since the majority of them is
also expressed at high transcript levels (FIGURE 2). The overall correlation between
transcript and relative protein abundance is weakly positive (Spearman Rank
correlation of 0.53) suggesting that the expression of nucleus encoded chloroplast
proteins is predominantly regulated at the transcriptional level (FIGURE 2).
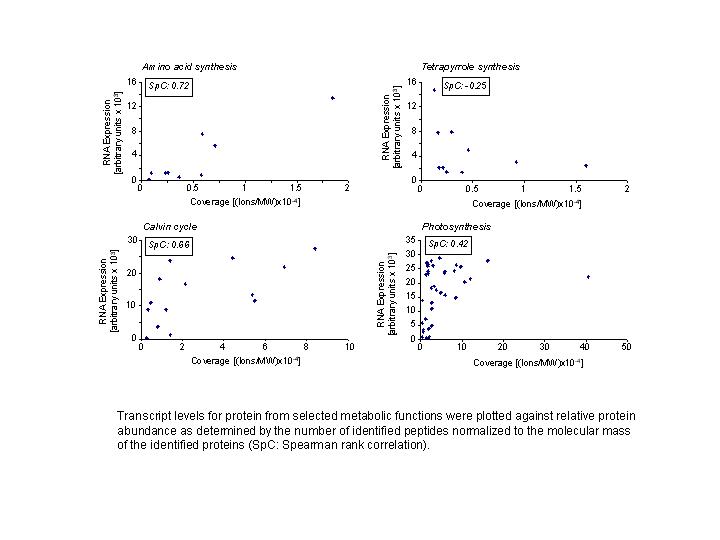
Interestingly, more detailed analyses suggested, that the correlation differs
between different biosynthetic pathways and metabolic functions (FIGURE 3). All
identified proteins can be searched in plprot,
either by key word or by BLAST search.
|
Top
|
Top
|
Top
|
|