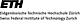plprot
- Plastids from red bell pepper (Capsicum annuum L.) fruits ["chromoplast"]

|
Further details in:
Siddique et al., Plant Cell Physiol., 2006 47: 1663-1673.
|
Bell pepper chromoplasts were isolated by Percoll density gradient centrifugation using a step
gradient developed with 10%-20%-30%-40%-50% and 60 % Percoll in GR buffer. After chromoplast
isolation we devised a serial fractionation procedure to extract and fractionate chromoplast
proteins on the basis of their differential solubility. Here, the proteins were fractionated
into soluble proteins (OSMO), peripheral membrane proteins (8M urea) and integral membrane
proteins (5% SDS) (FIGURE 1). The soluble proteins make up for 76 % and the peripheral
membrane proteins make up for 24 % of the total mass of proteins extracted (Fig. 1).
These values indicate that the isolated chromoplasts contain mostly soluble proteins
suggesting that the organelles were largely intact, because soluble proteins would
leak from defective organelles. Only one major protein was visible in the SDS fraction
upon silver staining (FIGURE 1). This protein was identified as capsanthin/capsorubin
synthase which is involved in the carotenoid biosynthesis pathway. The small number of
integral membrane proteins that were solubilized with the 5% SDS extraction step implies
that the chloroplast thylakoid membranes are disassembled and thylakoid membrane proteins
degraded during chromoplast formation. The latter represent the major protein mass in
chloroplasts and require SDS for their solubilization (Kleffmann et al., 2004).
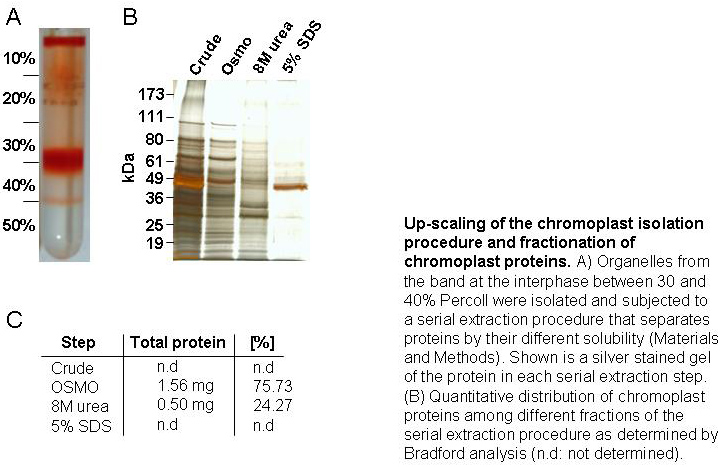
Because mass spectrometric identification of proteins depends on protein databases,
we expect that a database-dependent search strategy will not detect a significant
number of chromoplast proteins because of database constraints for bell pepper. In
order to assess how many peptides were not detected in the standard database search
and which proteins they identify, we devised an alternative data analysis strategy
to identify peptides in a database-independent fashion. This strategy comprises a
database-independent MS/MS spectrum quality scoring to identify peptide-derived
spectra that were not identified in a standard database search (Nesvizhskii et al., 2006).
Such high-quality spectra are subsequently subjected to de novo sequencing
(Frank and Pevzner, 2005). The de novo sequencing results are then filtered
on the basis of a reliability threshold before they are submitted to MS-BLAST
searches to identify peptides on the basis of homology (Shevchenko et al. 2001)
(FIGURE 2). From the 83 mass spectrometry runs that we performed in the course of
this study, 302430 MS/MS spectra were produced. In a standard database search using
SEQUEST and PeptideProphet, 4793 spectra were already identified with a confidence
of higher than 0.9. With the help of QUALSCORE, we identified 8666 high-quality
peptide spectra that were not identified in the database search.
Chromoplast development in bell pepper is accompanied by the massive synthesis and
accumulation of carotenoids. Most of these pigments (>95%) accumulate in lipoprotein
fibrils which are supramolecular structures that contain carotenoids, some lipids and
fibrillin, a protein that is the main structural component in these assemblies.
Fibrillin (annotated as plastoglubule associated protein) is one of the most abundant
proteins in bell pepper chromoplasts and we detected 210 peptides in different protein
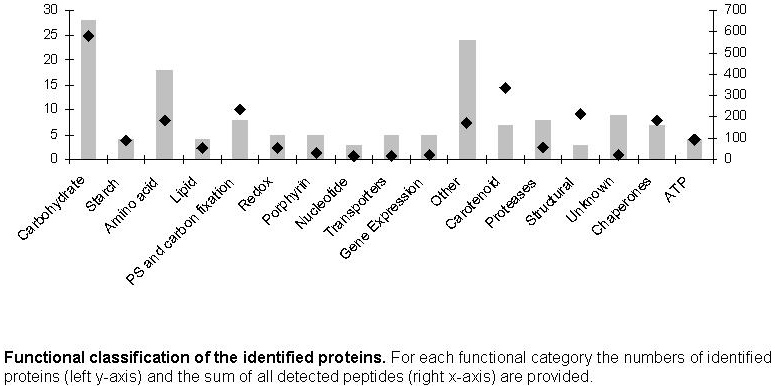
fractions (FIGURE 3). We identified the carotenoid biosynthetic pathway as the
predominant individual pathway and achieved coverage greater than 50% (Fig. 3).
Capsanthin/capsorubin synthase (CCS) is the most abundant chromoplast enzyme with
293 detected peptides (FIGURE 3). Most of the other identified proteins have a
general function in the cellular metabolism, and the majority (25 proteins, 580
detected peptides) is involved in the carbohydrate metabolism (FIGURE 3). We have
identified transketolase, transaldolase, chloroplast phosphoglycerate kinase,
glyceraldehyde-3-phosphate dehydrogenase and triosephosphate isomerase with many
peptides signifying their high abundance). Information about all identified proteins
is deposited in plprot, that can be searched by either key word or by BLAST search.
|
Top
|
Top
|
Top
|
|