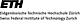plprot
- From Etioplasts to Chloroplasts: Proteome dynamics during plastid differentiation in rice (Oryza sativa)

|
Further details in:
Kleffmann et al.,Plant Physiol, 2007 143: 912-923.
|
Although etioplasts represent an end product of plastid differentiation in the dark,
they can rapidly initiate the re-differentiation to chloroplasts when light is perceived.
We have used this experimental system to analyse the molecular mechanisms that initiate
chloroplast differentiation from etioplasts. To this end, we started out by mapping the
rice etioplast proteome by 2-dimensional gel electrophoresis. The etioplast proteome
map provides insights into the abundance of the identified proteins and helps to infer
the prevalence of different metabolic pathways that constitute the etioplast metabolism.
This is feasible since the Sypro Ruby stain that we have used for visualization is a
proportional stain that allows to directly correlating staining intensity with protein
abundance. A comprehensive depiction of the total protein mass distribution to specific
metabolic functions in rice etioplasts is presented in Figure 1. The highest number of
identified proteins (diamonds, upper graph) and the majority of the total protein mass
(gray bars, upper graph) participate in amino acid and carbohydrate metabolism. In
addition, a significant fraction of the total protein mass is allocated to gene
expression, secondary metabolism and tetrapyrrole biosynthesis (FIGURE 1). Proteins
from the nucleotide and fatty acid metabolism are of minor account in the etioplast
proteome (FIGURE 1).
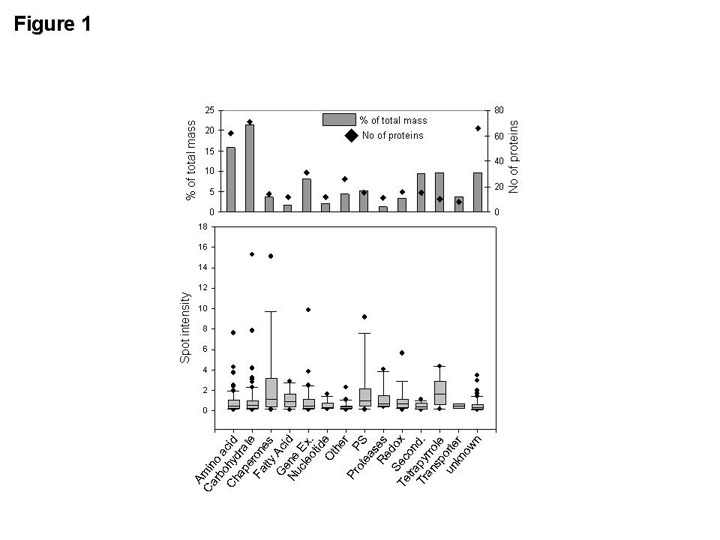
On the basis of the quantitative protein information inferred from the 2 D map, we
analysed the early light-induced changes of the etioplast proteome and provide here
a comprehensive sketch of the global re-organization of the heterotrophic etioplast
metabolism to the autotrophic chloroplast metabolism (FIGURE 2). More precisely, we
analysed the protein abundance after 2 hours, 4 hours and 8 hours of illumination
and compared these values with the 0 hour control (etioplast). During this time,
etioplasts initiate the synthesis of chlorophyll and the assembly of a functional
thylakoid membrane system. We observe a transition of the etioplasts metabolism
towards photosynthesis-dependent autotrophy, and find at the regulatory level,
that nuclear-encoded plastid RNA binding proteins are phosphorylated in a light
dependent manner, i.e. the degree of phosphorylation increases with illumination
time. This is a clear indication that plastid mRNA stability is one of the major
regulatory switches for the initiation of light-induced chloroplast development.
All identified proteins together with their abundances and their regulation by
light can be searched in plprot, either by key word or by BLAST search.
|
Top
|
Top
|
|